|
|
Biocomputation Research The scope of our Center is broad enough to encompass many biomedical research domains, and yet is focused in its mission to use physical and mathematical modeling to create accurate simulations of biological structure and function. In analyzing the driving biological problems, we have found abundant commonalities to justify the development of a common simulation platform. Data, model, and simulation (and their relationships) provide the basis for a general framework for conducting biophysical investigations across a range of scientific domains and physical scales. (Fig. 1.1)
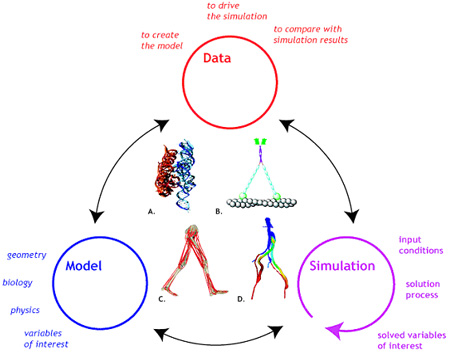
Figure 1.1. We enable physics-based modeling and simulation of biological structures, including (A) RNA folding, (B) myosin dynamics, (C) Protein Folding (D) neuromuscular dynamics, and (E) cardiovascular dynamics. Data describing atomic structures, anatomical geometry, and other parameters are used to build models. A model describes the geometry, biology, and physics of the system through governing equations. A simulation is produced by integration of the governing equations. Simulations are compared to data describing the performance of the system to test the accuracy of the functional predictions. This modeling and simulation framework provides a powerful paradigm for approaching biological problems with a structural perspective across a broad range of scales, from atoms to organisms. The Simulation Tool Kit (SimTK) is available at Simtk.org and can be downloaded from Simtk.org/home/simtkcore.
In addition to integrating existing methods for modeling, simulation, and data analysis, we will develop new mathematical and computational tools to advance the field of physics-based simulation. Many computational research challenges arise at all scales, including the need to:
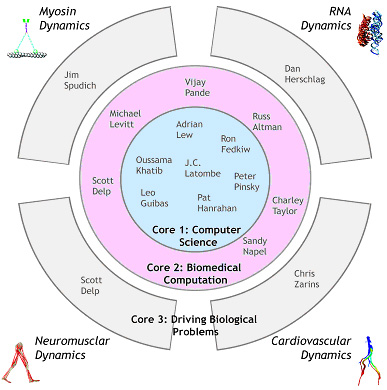
There also are fundamental differences in simulating physics at different scales that must be acknowledged. These do not make integration impossible, but instead suggest that some research challenges will be most important only at certain scales. For example, electrostatic forces are dominant at the molecular level, while modeling inertial forces is critical at the macroscopic level. Our team includes computer scientists and biomedical computational scientists (Fig. 1.3). Core 1 is composed of applied mathematicians, computer scientists and engineers who are interested in fundamental algorithm development. Core 2 is composed of biomedical computation specialists who live in two worlds: they speak the language of their biological or medical domain, and they understand and can implement the algorithmic innovations from Core 1. They can also translate the needs of the biologists (Core 3) into terms that the Core 1 scientists can understand. The Simbios Publication list show many examples of these interactions. Our continuuing efforts have led to routine interactions between faculty involved in molecular simulation and those involved in biomechanical simulation. We were struck that the principles used to model and simulate in these very different application areas are not only similar, but can be brought under a single rubric with sufficient generalization of terminology. Figure 1.3. Investigators in Core 1 focus on basic data structure and algorithm development for modeling and simulation. Investigators in Core 2 focus on using these data structures and algorithms to create tools and techniques for modeling across scales. Our driving biological problems (DBPs) create requirements for the tools and techniques, and evolve to continually stress the capabilities of SimTK. |
|